Configuring R Environments
Once you have obtained the repository URL, your environment must be configured to use the URL to download and install packages.
Base R
To configure R to use Package Manager as its CRAN repository, set the repos option to use the repository URL:
R
options(repos = c(CRAN = "https://packagemanager.posit.co/cran/__linux__/rhel9/latest"))Alternatively, instead of replacing your default URL, additional repositories can be added to your existing configuration. For example:
R
local({
repos <- c(PackageManager = "https://packagemanager.posit.co/cran/__linux__/rhel9/latest")
repos["LocalPackages"] <- "https://packagemanager.posit.co/local/__linux__/rhel9/latest"
# add the new repositories first, but keep the existing ones
options(repos = c(repos, getOption("repos")))
})
# verify the current repository list
getOption("repos")Result:
PackageManager
"https://packagemanager.posit.co/cran/__linux__/rhel9/latest"
LocalPackages
"https://packagemanager.posit.co/local/__linux__/rhel9/latest"
CRAN
"https://cloud.r-project.org"The same code can be added to your R startup file (~/.Rprofile) to maintain the repository configuration across R sessions. See our article on managing R startup files for more information.
If your repository requires authentication, follow the additional steps to configure R to use authenticated repositories.
If you are installing packages on Linux and want to take advantage of pre-built binary packages for speed and reliability, follow the instructions in Configuring the R User Agent Header to set up or verify your configuration.
RStudio IDE
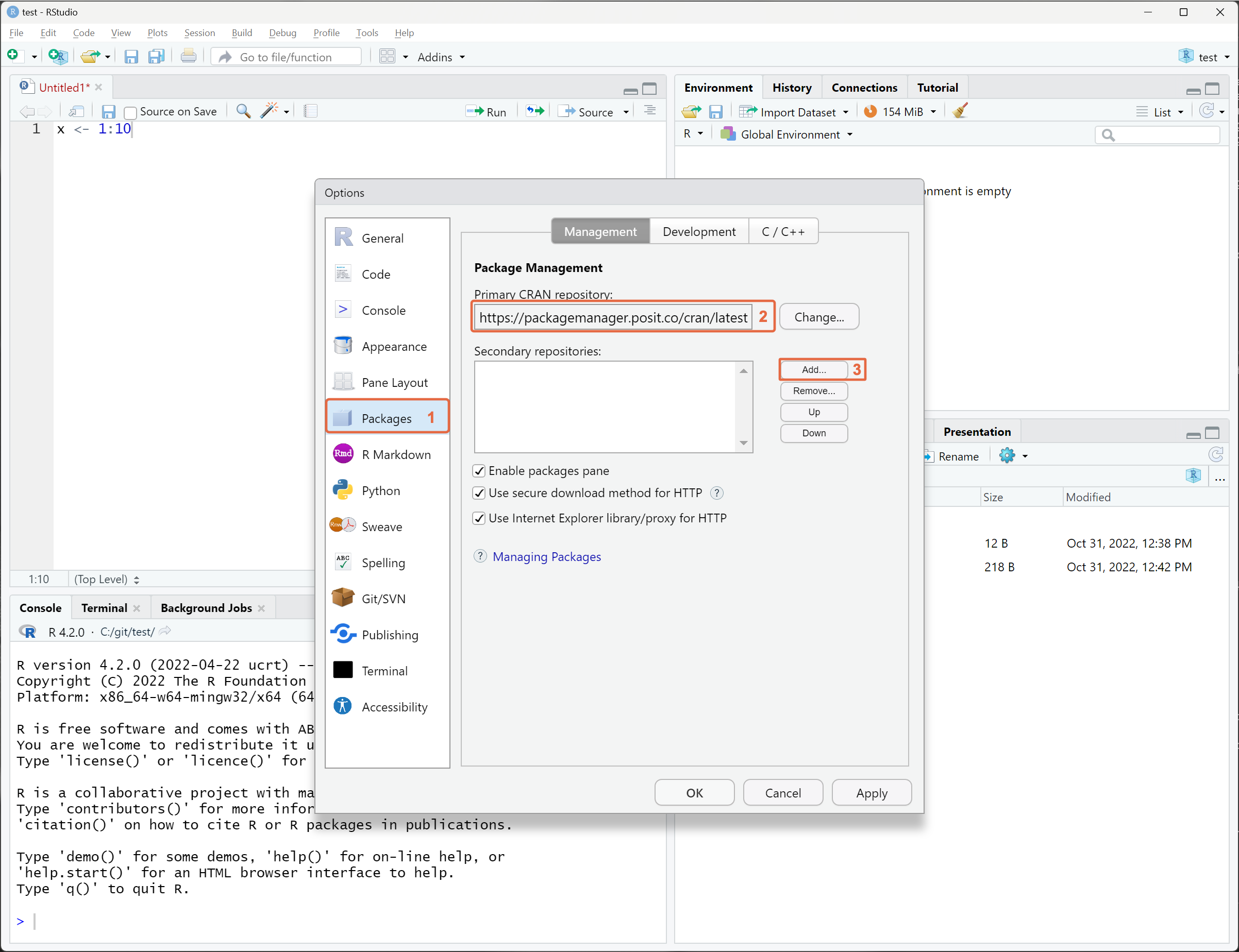
In the RStudio IDE, repository settings are configured through the Global Options menu available from the Tools section of the main toolbar.
If your repository requires authentication, follow the steps to configure R to use authenticated repositories before proceeding.
From the left pane, click Packages 1, paste the repository URL into the Primary CRAN repository field 2 or click Add 3 to include this repository as a secondary repository.
If you are using RStudio from Posit Workbench, your administrator can also configure the default repository for all users. See Configuring Posit Workbench for more information.
Positron
In Positron, repository settings are configured in the same way as configuring base R. Please refer to the instructions for configuring base R.
If you are using Positron from Posit Workbench, your administrator can also configure the default repository for all users. See Configuring Posit Workbench for more information.
Authenticated repositories
If your Package Manager repository requires authentication, you can configure R to use a .netrc file for credentials and curl as the download method.
This method is widely supported and provides a secure way to access authenticated repositories in R. It can be used with various tools and IDEs, including install.packages(), devtools, rsconnect, renv, BiocManager, pak, RStudio, and Positron.
Some tools, such as renv and BiocManager, require additional configuration to use the .netrc file with curl. Others, like pak, support alternative authentication methods, such as using the system keyring service.
Getting a token
Your Package Manager administrator can provide you with a token to use for authentication. Refer to Creating API Tokens for details.
Configure R to use .netrc and curl
Create a
.netrcfile in your home directory at~/.netrc.Add the following content, replacing
[packagemanager.example.com]with your Package Manager server address and[your-token]with your actual token.~/.netrc
machine [packagemanager.example.com] login __token__ password [your-token]The machine field should not contain a protocol, port, or repository path. If your repository URL is https://packagemanager.example.com/cran/latest, use packagemanager.example.com for the machine field.
Since the credentials are stored in plaintext, ensure that the
~/.netrcfile is only accessible to you by running:Terminal
chmod 600 ~/.netrcConfigure R to use
curlas the download method by adding the following lines to your R startup file (~/.Rprofile).~/.Rprofile
# Use curl with a netrc file for authenticated repo access options(download.file.method = "curl") options(download.file.extra = paste( "--netrc", # Follow redirects, show errors, and display the HTTP status and URL '-fsSL -w "%{stderr}curl: HTTP %{http_code} %{url_effective}\n"', # Configure the R user agent header to install Linux binary packages sprintf('--header "User-Agent: R (%s)"', paste(getRversion(), R.version["platform"], R.version["arch"], R.version["os"])) ))You will need additional configuration to install Linux binary packages with
curl, so that is also included here.
Create a
.netrcfile in your home directory at~/.netrc.Add the following content, replacing
[packagemanager.example.com]with your Package Manager server address and[your-token]with your actual token.~/.netrc
machine [packagemanager.example.com] login __token__ password [your-token]The machine field should not contain a protocol, port, or repository path. If your repository URL is https://packagemanager.example.com/cran/latest, use packagemanager.example.com for the machine field.
Since the credentials are stored in plaintext, ensure that the
~/.netrcfile is only accessible to you by running:Terminal
chmod 600 ~/.netrcConfigure R to use
curlas the download method by adding the following lines to your R startup file (~/.Rprofile).~/.Rprofile
# Use curl with a netrc file for authenticated repo access options(download.file.method = "curl") options(download.file.extra = paste( "--netrc", # Follow redirects, show errors, and display the HTTP status and URL '-fsSL -w "%{stderr}curl: HTTP %{http_code} %{url_effective}\n"' ))
Create a
.netrcfile in your home directory at~/.netrc. This may be your Documents folder (typicallyC:\Users\username\Documents). You can find the location by runningpath.expand("~/.netrc")in an R console.Add the following content, replacing
[packagemanager.example.com]with your Package Manager server address and[your-token]with your actual token.~/.netrc
machine [packagemanager.example.com] login __token__ password [your-token]The machine field should not contain a protocol, port, or repository path. If your repository URL is https://packagemanager.example.com/cran/latest, use packagemanager.example.com for the machine field.
Configure R to use
curlas the download method by adding the following lines to your R startup file (~/.Rprofile).~/.Rprofile
# Use curl with a netrc file for authenticated repo access options(download.file.method = "curl") options(download.file.extra = paste( "--netrc", # Follow redirects, show errors, and display the HTTP status and URL '-fsSL -w "%{stderr}curl: HTTP %{http_code} %{url_effective}\n"' ))
.netrc files only support configuring a single set of credentials per server. This means that if you have multiple repositories, you need to use the same token for all of them. The token must have access to each repository.
Additional configuration for renv, BiocManager, and pak
You can configure renv to use credentials from .netrc by adding the following line to your R startup file (~/.Rprofile).
~/.Rprofile
# Configure renv to install from authenticated repos
Sys.setenv(RENV_DOWNLOAD_METHOD = "curl")To install packages from an authenticated Bioconductor repository, ensure you have BiocManager version 1.30.24 or later.
Once updated, follow the Setup page instructions in the Package Manager UI to configure your Bioconductor mirror in R.
pak supports authenticated repositories via a .netrc file or the system keyring service, but only in its latest development version.
If you are exclusively using pak for package installation, we recommend using the system keyring as a more secure option for storing credentials than .netrc. However, this method may require user interaction and may not work in all scenarios.
For details, refer to the pak documentation on Configuring authenticated repositories. For instructions on installing the development version of pak, refer to the pak documentation on Nightly builds.
Checking for Success
If you changed any R startup files (e.g., ~/.Rprofile), restart your R session to apply the changes.
For CRAN or internal package repositories, running getOption("repos") in R should show the URL of the Package Manager instance. For Bioconductor repositories, running getOption("BioC_mirror") should also show the URL of the Package Manager instance.
For a complete test, you can also confirm that packages are installed from the Package Manager instance by default. To test a CRAN or internal package repository, run the following commands to install a package in the repository:
R
# Show the configured repos
getOption("repos")
# Install a package in the repository, such as A3
install.packages("A3")While you can install any package as a test, one that is small and has few dependencies will be the quickest. The A3 package on CRAN is the first alphabetically that meets this criteria.
To test the Bioconductor repository, you can download and install a Bioconductor package:
R
# Show the configured Bioconductor repo
getOption("BioC_mirror")
# Install the BiocManager package from CRAN
install.packages("BiocManager")
# Check that BiocManager uses Package Manager for the Bioconductor repositories
BiocManager::repositories()
# Install a Bioconductor package
BiocManager::install("BiocVersion", update = FALSE)